Sharp Laboratory : Structural Mass Spectrometry
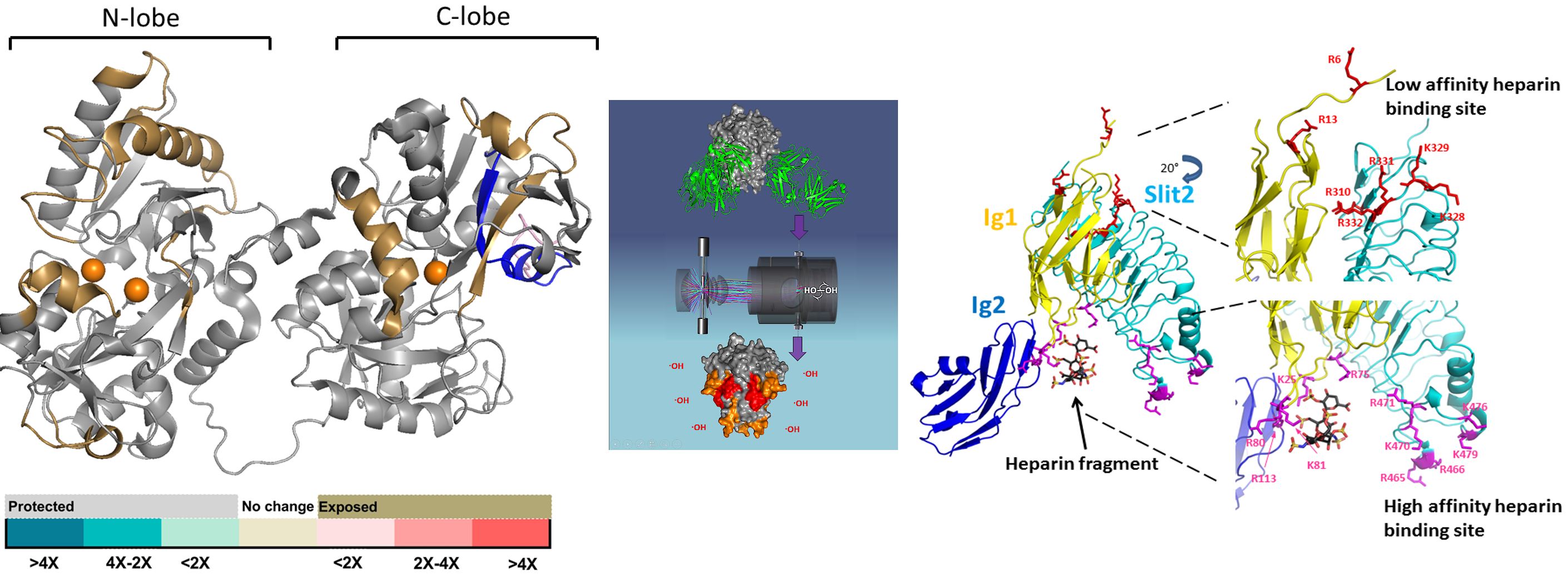
Prof. Sharp’s group is active in the development and application of new mass spectrometry-based technologies for studying the structure-function relationships of proteins and carbohydrates of biomedical interest. Structural biology is key to understanding the molecular basis of biology and medicine. While modern structural biology tools are incredibly powerful, there remains significant categories of biomolecular systems that are challenging or impossible to properly characterize using existing high-resolution technologies such as dynamic systems, systems in a complex or heterogeneous matrix, and protein:carbohydrate complexes. The Sharp Lab develops new technologies that use protein and carbohydrate chemistry coupled with modern analytical technologies to characterize the structure of such systems.
NEWS